Enabling large-scale testing of cancer drugs with machine learning
Prof. Euisik Yoon and his team developed a new machine learning tool that enables large-scale testing of cancer drug effectiveness with microfluidics.
 Enlarge
Enlarge
Prof. Euisik Yoon and his team of researchers have developed a new tool to improve drug screening for common cancer treatment drugs. They designed a machine learning program that makes large-scale drug screening efficient and low cost for use with microfluidic platforms, which yield far more accurate results than traditional drug screening.
Traditional testing of cancer drugs begins in a 2D monolayer cell culture, which is cheaper and faster than animal testing. Cancer cells are placed on substrate, treated by drugs, and then reexamined to determine the effectiveness of the specific drug. This approach cannot replicate in vivo (conditions found within a living organism) microenvironments. On the contrary, 3D cell culture allows cancer cells to form tumor spheres in suspension or hydrogel, which better mimics the profile of drug exposure, nutrients, and oxygen supplies in real tumors.
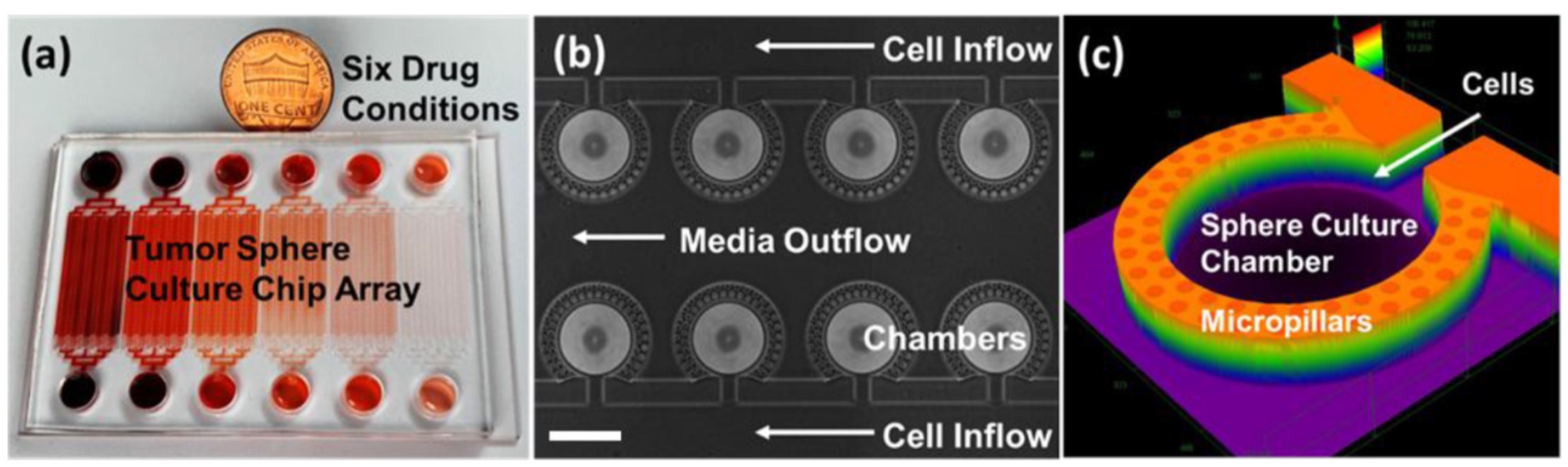
Microfluidic technology works well with 3D cell growth by allowing researchers to manipulate the cancer cells and control the size and environment for 3D cell growth. In addition, thousands of microchambers can be realized on a chip, allowing for high-throughput screening.
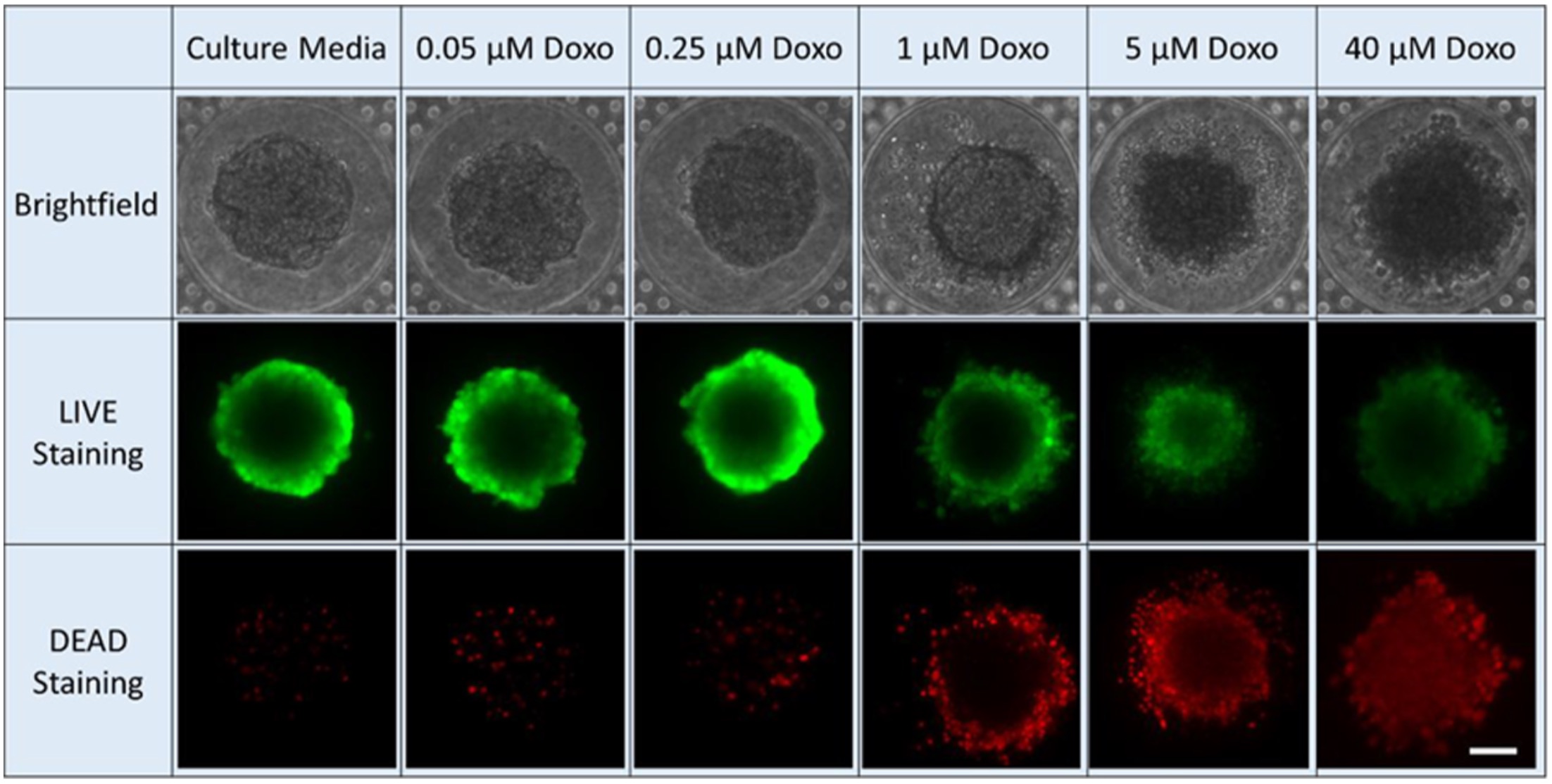
However, using microfluidic technology to test drugs is far more expensive and slower than other methods. To overcome these setbacks, Yoon and his team trained a convolutional neural network that automatically collects images, crops them, and analyzes them. This network collects bright-field images (a microscopy technique where the sample is illuminated against a bright background) and their subsequent drug-inhibition scores. Drug inhibition scores are grounded on conventional LIVE/DEAD staining and fluorescence imaging, where the cancer cells are “stained” to mark the viability of individual cells. This technique is typically slow and only provides an endpoint analysis.
“Automation and efficient readout methods are the key for microfluidics to get to the next level in the next decade,” says Yu-Chih Chen, Assistant Research Scientist in ECE who helped lead the project.
But machine learning only works if you’re training it with information provided by direct observation, which is why Yoon says it was important for his students to have some background or understanding of biology.
“Domain knowledge is really, really important when you apply A.I.,” Yoon says. “Otherwise, you cannot draw meaningful conclusions from this.”
 Enlarge
Enlarge
The team’s machine learning model was able to accurately judge the drug inhibition score of a tumor sphere using only the bright-field image. They used this method to precisely estimate drug efficacy of three chemotherapy drugs: doxorubicin, oxaliplatin, and irinotecan. What’s more, they found common bright-field morphological features that indicate sphere viability across different drugs. This suggests it’s possible to train a generic model using a few representative drugs and apply it to many different drugs in large-scale, efficient, and low-cost screening.
ECE doctoral students Zhixiong Zhang and Lili Chen were co-first authors on the paper. The work of coding the machine learning program was done by undergraduate students Lili Chen and Tiantian Zhang, who participated in the research as part of the Summer Undergraduate Research in Engineering (SURE) program.
“With these sorts of projects, undergraduates get to apply their knowledge to real-life problems,” Yoon says. “And, also, when we’re working with young, smart, undergrad students, sometimes they do better than we do. ‘We’ means the faculty.”
The paper, “Label-Free Estimation of Therapeutic Efficacy on 3D Cancer Spheres Using Convolutional Neural Network Image Analysis,” by Zhixiong Zhang, Lili Chen, Yimin Wang, Tiantian Zhang, Yu-Chih Chen, and Euisik Yoon, is available here.
The work was supported by the National Institutes of Health, the University of Michigan Office of Research, and the Forbes Institute for Cancer Discovery. The device was fabricated in the University of Michigan Lurie Nanofabrication Facility.
 MENU
MENU 
