New computer vision technique enhances microscopy image analysis for improved cancer diagnosis
A team of researchers from University of Michigan and Michigan Medicine has collaborated to develop a new computer vision learning technique for cancer diagnosis. Their tool, HiDisc, uses artificial intelligence and machine learning to analyze microscopy images and identify common features of cancerous tumors.
Made up of researchers from Computer Science and Engineering, the Department of Computational Medicine and Bioinformatics, and the Department of Neurosurgery, the team will present their research at the upcoming IEEE / CVF Computer Vision and Pattern Recognition Conference (CVPR), the premier global event in computer vision, taking place June 18-22, 2023, in Vancouver, Canada. Their paper, Hierarchical Discriminative (HiDisc) Learning Improves Visual Representations of Biomedical Microscopy, is one of a select group to be chosen as a “Highlight” at the conference.
HiDisc uses advanced AI techniques to make the process of analyzing biomedical microscopy images both more efficient and more accurate. Cheng Jiang, PhD candidate in computational medicine and bioinformatics and lead author on the project, emphasizes the importance of this development, highlighting the lengthy process traditionally involved in cancer diagnosis.
“The conventional workflow is that a specimen from a tumor is sent to the pathology lab, where it can take a long time for the tissue to be analyzed,” Jiang describes. “Nowadays, we have a new type of high-resolution optical microscope, which allows for much quicker imaging of tumor specimens, but this still requires a pathologist to look at the images to make a diagnosis.”
The goal of HiDisc is to make it much easier and faster for physicians to make a diagnosis, taking a matter of only minutes, rather than days, to analyze and classify microscopy images. “Essentially,” Jiang says, “our goal is to use AI to help clinicians make a diagnosis based on these tumor images virtually instantaneously during surgery, while the surgeon is waiting.”
One of HiDisc’s primary innovations is its ability to analyze multiple patches from the same tumor as a whole, rather than individually. This allows the model to better learn and characterize entire tumors, instead of treating smaller images of the same tumor separately, which can create redundancies in the analysis.
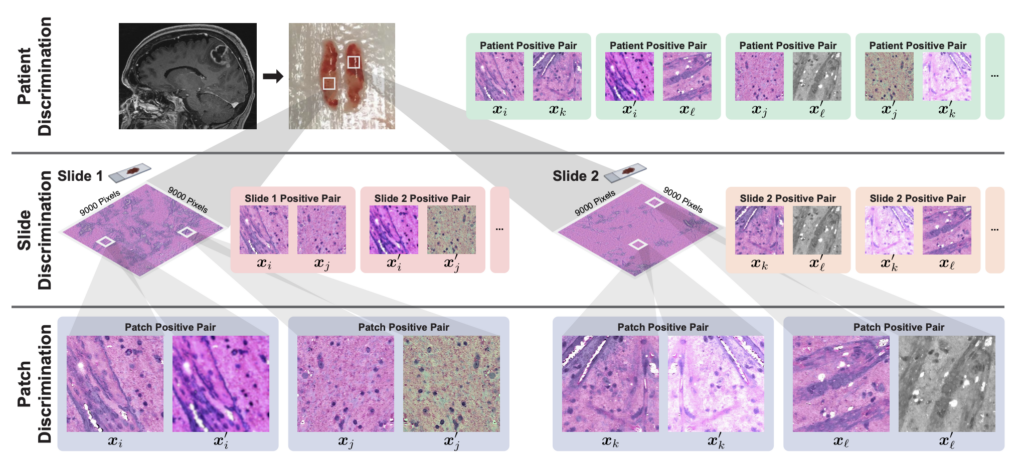
“Images collected via biomedical microscopy are very, very large. These aren’t your typical 500 by 500 pixel images; they can be up to a billion pixels,” Jiang says. “Current practice is that these images are divided into smaller patches and analyzed independently.”
Considering each patch separately is not ideal, however, and introduces significant inefficiencies in the machine learning process. “This approach assumes that each patch is independent, even though they’re from the same tumor,” says Jiang. “This results in a much lower learning efficiency, because we’re learning to differentiate views of the same underlying biology.”
HiDisc resolves this issue by leveraging the inherent data structure of biomedical microscopy to label and group patches from the same tumor. The tool encodes different views of the same tumor with similar representations so that patches are considered collectively rather than separately.
HiDisc is also unique in that it reduces reliance on data augmentations, a method commonly used to corrupt images to allow AI to learn the difference between tumors. “These augmentations,” says Jiang, “risk erasing information that could be relevant to making a diagnosis.”
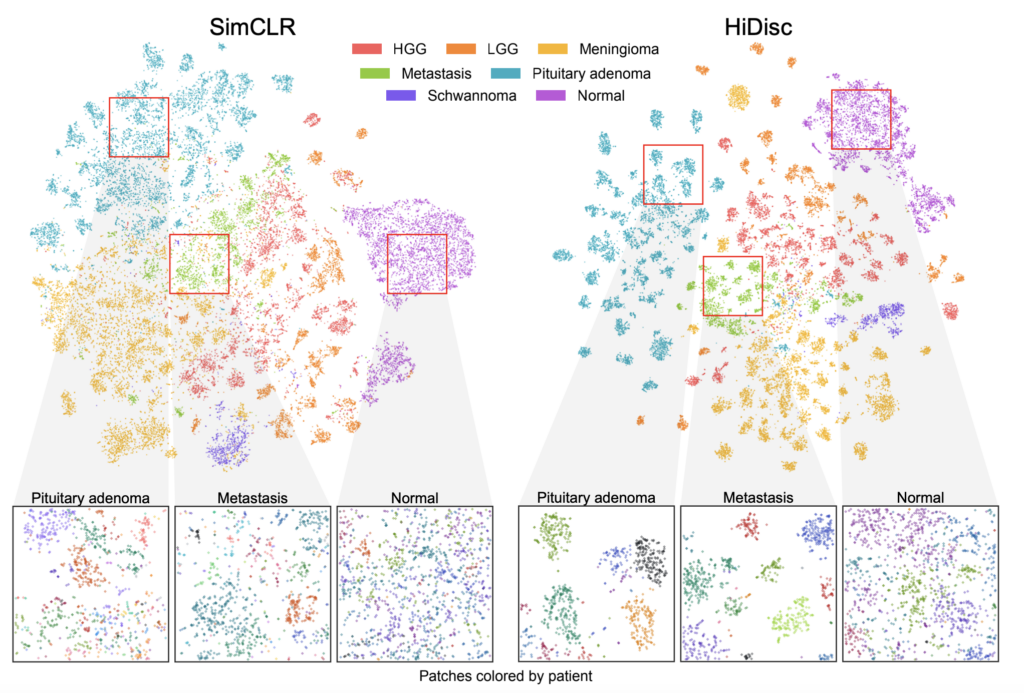
Because it does not rely on data augmentations to transform and label images like most existing methods, HiDisc is a more natural and accurate way of correlating multiple patches from the same tumor. By allowing clinicians to understand and analyze images on a tumor level, rather than based on isolated patches, HiDisc provides them with a stronger, faster, and more holistic diagnostic tool.
Jiang and his coauthors validated HiDisc against several other state-of-the-art self-supervised learning methods for cancer diagnosis, demonstrating that HiDisc substantially outperforms other approaches in all metrics. Compared to the best-performing baseline method, HiDisc improves image classification by nearly 4%, reaching close to 88% accuracy in certain cases.
The team conducted their validations using microscopy images of brain tumors, and Jiang emphasizes that HiDisc “can be generalized to any type of tumor using any type of microscopy.” Jiang and his colleagues are currently working on further aggregating patch representations learned using their method into patient-level representations, which will enable even more accurate and comprehensive image analysis and classification. The ultimate goal, he says, is “to implement this directly in the operating room, so we can use AI to make surgery and diagnosis easier and faster.”
 MENU
MENU 
